Overview
In graduate school, my research focused on understanding how human cells maintain genome stability by mapping DNA repair events in single cells. Specifically, I studied the RecQ family of DNA helicases (including RECQL5, BLM, WRN, and RECQL1), a group of genome “caretaker” enzymes linked to aging and cancer biology. My PhD work used single cell template strand sequencing (Strand-seq) to pinpoint where repair-driven genome alterations occur across the genome, at kilobase-scale resolution, far beyond what classic cytogenetic assays can localize.
Research focus and approach
I built a large panel of CRISPR-Cas9 single and double knockouts in the haploid KBM7 model system (over 20 genotypes) to systematically test how loss of specific helicases changes patterns of genome instability. Using Strand-seq, I mapped sister chromatid exchange (SCE) events and copy number alteration (CNA) breakpoints at kilobase resolution and compared how different helicase-deficient backgrounds reshape these repair signatures.
Key findings
A major biological insight from this work is that repair events are not randomly distributed. SCEs were frequently enriched near actively transcribed genes, and near genomic elements such as G-quadruplex (G4) motifs and common fragile sites, supporting a model where RecQ helicases protect the genome at regions that are challenging for faithful replication and repair.
Method development and impact
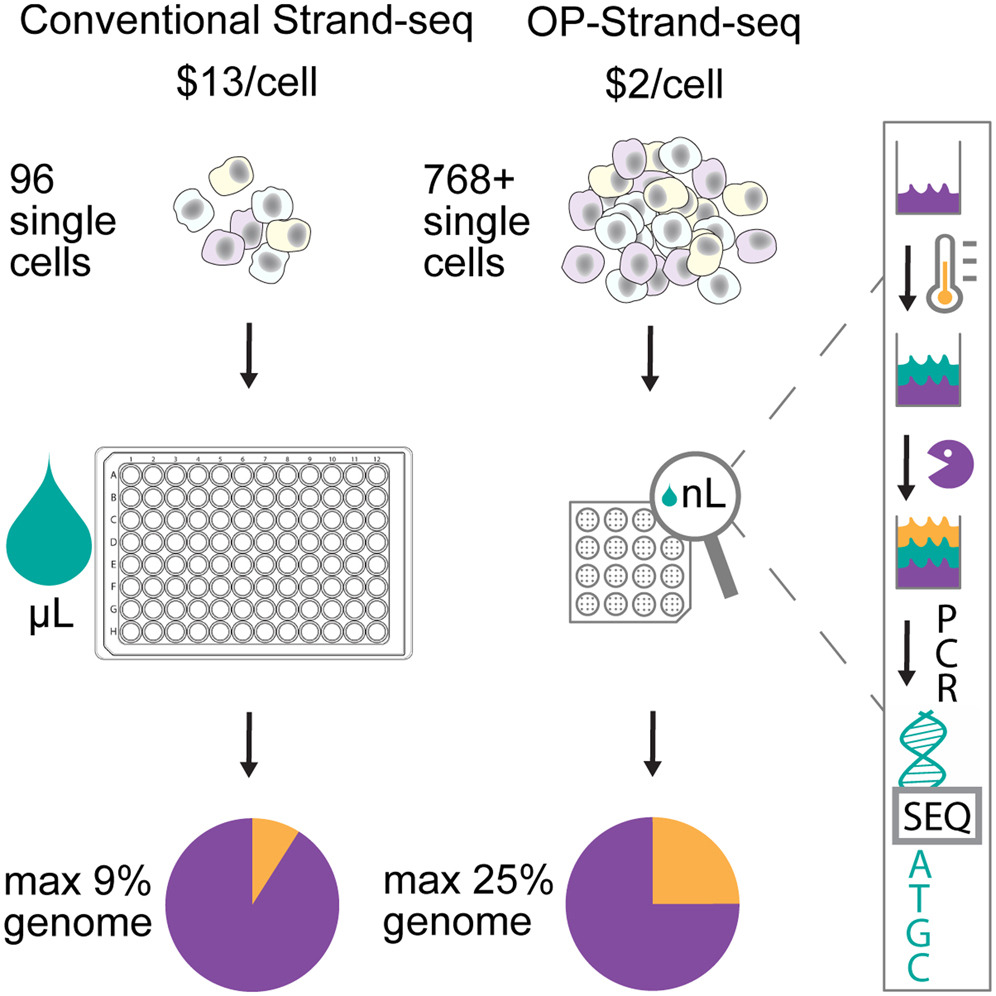
I contributed to improving the scalability of Strand-seq through a one-pot, nanoliter-volume library prep approach (OP-Strand-seq), enabling high-throughput single-cell sequencing at low cost while capturing ~10% to 25% of the genome per cell. This approach reduced per-library costs to roughly US$1 to US$2 and increased throughput (hundreds of cells per run), making large-scale single-cell directional genomics more accessible. Beyond SCE mapping, I also developed bioinformatic strategies to generate genotype-specific callsets for CNAs, inversions, and translocations, expanding the utility of Strand-seq for structural variant discovery and genome instability research.
Significance
Together, this body of work helped connect helicase function to specific, vulnerable genomic contexts and strengthened Strand-seq as a practical platform for studying DNA repair, recombination, and structural variation in single cells, with clear relevance to mechanisms underlying cancer predisposition and aging.